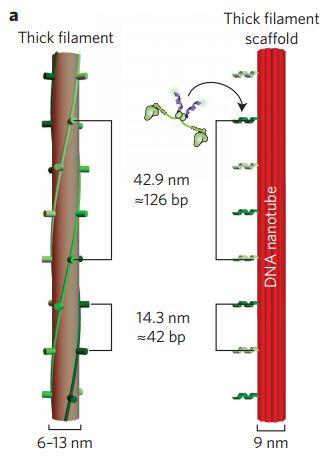
One of the challenges in biology is the ability and manipulate and study large multi-protein ensembles, such as signaling complexes and vesicle transport machinery. While in vivo approaches have provided significant information on organization and composition of large macromolecular complexes, the cellular environment is too complex for detailed molecular analysis. Further, these complexes are often weakly-coupled and therefore not amenable to standard biochemical and structural approaches. One approach is to use DNA nanotechnology to synthetically pattern multi-protein ensembles. During my postdoc, I have been developing new tools for reconstituting and studying the molecular properties of both multi-motor complexes (Fig 1) and macromolecular signaling complexes.
Publications
(1) Hariadi RF*, Sommese RF*, Adhikari AS, Taylor RE, Sutton S, Spudich JA, Sivaramakrishnan S. Mechanical coordination in motor ensembles revealed using engineered artificial myosin filaments. 2015 Nature Nano 10:696-700. * Authors contributed equally
(2) Hariadi RF, Sommese RF, Sivaramakrishnan S. Tuning myosin-driven sorting on cellular actin networks. 2015 eLife. doi: 10.7554/eLife.05472.
(3) Sommese RF, Hariadi RF, Kim K, Liu M, Tyska MJ, Sivaramakrishnan S. Patterning Protein Complexes on DNA Nanostructures Using a GFP-Nanobody. 2016 Protein Science 25(11):2089-2094.