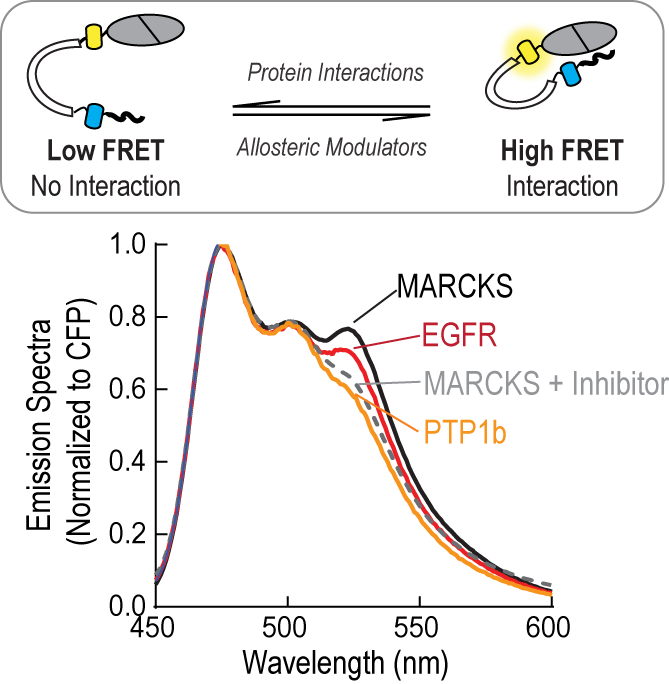
A critical question in biology is how specificity is encoded in the dynamic protein interactions that occur within the complex and crowded cellular environment. This becomes particularly interesting when one considers signaling molecules like kinases and phosphatases which not only couple diverse stimuli to distinct cellular processes but must also interact with a variety of substrates and regulatory proteins. During my postdoc, I have created a series of FRET-based tools to dissect the molecular mechanisms driving conformational regulation and substrate specificity in kinases. These approaches are now providing new tools for dissecting and therapeutically targeting the functional properties of signaling molecules like Protein Kinase C
Publications
(4) Sommese RF, Sivaramakrishnan. Substrate Affinity Differentially Influences Protein Kinase C Regulation and Inhibitor Potency. 2016 J Biol Chem 291(42):21963-21970.
(5) Swanson CJ, Sommese RF, Petersen KJ, Ritt M, Karslake J, Thomas DD, Sivaramakrishnan S. Calcium stimulates self-assembly of Protein Kinase C α in vitro. 2016 PLoS One 11(10):e0162331.
(6) Sommese RF, Ritt M, Swanson CJ, Sivaramakrishnan S. The role of regulatory domains in maintaining auto-inhibition in the multi-domain kinase PKCα. J Biol Chem Epub Date (TBA)