Mapping the multi-domain regulation of cell signaling using the Kinase Toolbox
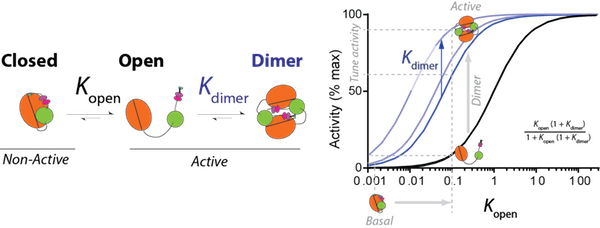
Kinase mediated phosphorylation of proteins broadly regulates cellular responses in normal and disease states. Kinases in cell signaling networks function as ‘micro-processors’ that couple different stimuli to distinct outputs. They do so by regulating intra- and inter-molecular protein interactions to influence the kinase catalytic cycle. However, contemporary methods to probe their function treat them primarily as binary switches that are simply activated or inactivated by stimuli. Therefore, we are limited in our ability to understand and control the versatility and specificity of kinase function. We have developed a toolbox of FRET-based sensors to map inter-domain interactions in protein kinases (Swanson and Sivaramakrishnan, JBC 2014). Our published studies with focal adhesion kinase (FAK; Ritt et al., JBC, 2013) and protein kinase C (PKC; Swanson et al., JBC, 2014) have uncovered new mechanisms of regulation of these kinases (Fig. 1). Ongoing studies reveal that in multiple AGC kinases, intra- and inter-domain interactions are sensitive to conformational changes initiated by nucleotides or inhibitors bound to the catalytic site. We are currently focused leveraging the kinase toolbox to identify isoform-specific kinase inhibitors
Selected Publications
- Kinase inhibitors allosterically disrupt a regulatory interaction to enhance PKCα membrane translocation. Lippert L*, Ma N*, Ritt M*, Jain A, Vaidehi N, Sivaramakrishnan S. J biol Chem. 2021 Jan 25;doi: 10.1016/j.jbc.2021.100339
Full text: Link Coming soon
- Bitopic Inhibition of ATP and Substrate Binding in Ser/Thr Kinases through a Conserved Allosteric Mechanism. Ma N, Lippert L, Devamani T, Levy B, Lee S, Sandhu M, Vaidehi N, Sivaramakrishnan S. Biochem. 2018 Oct 18;57(45): 6387–90
- Distinct structural mechanisms determine substrate affinity and kinase activity of Protein Kinase Cα. Lee S, Devamani T, Song HD, Sandhu M, Larsen A, Sommese R, Jain A, Vaidehi N, Sivaramakrishnan S. J Biol Chem. 2017 Sep 29;292(39):16300-16309
- Substrate Affinity Differentially Influences Protein Kinase C Regulation and Inhibitor Potency. Sommese RF, Sivaramakrishnan S. J Biol Chem. 2016 Oct; 291(42):21963-21970
- Conserved Modular Domains Team Up to Latch-Open Active PKCα. Swanson CJ, Ritt M, Wang W, Lang MJ, Narayan A, Tesmer JJ, Westfall M, Sivaramakrishnan S. J Biol Chem. 2014 Jun 20;289(25):17812-29