Cellular communication or signaling employs vast interconnected networks of proteins, with varying patterns of transient interactions that translate diverse extracellular stimuli to distinct cellular responses. While much is known about the structure and function of the individual components of signaling networks, a systems understanding of the decision-making process employed by the cell and its spatio-temporal deployment is still in its infancy. Our lab at the University of Minnesota is focused on engineering protein interactions in vitro, in live cells, and in whole organisms, in order to bridge the gap between our structural understanding of proteins and their emergent cellular function. Research in the lab targets four layers of information that are subtly embedded in any protein interaction cascade namely, protein conformation, partner selection, coordination of multi-domain interactions, and communication in multi-protein complexes (Fig. 1). We combine DNA nanotechnology (Hariadi et al., Nature Nanotechnology, 2015) and synthetic protein engineering (Sivaramakrishnan and Spudich, PNAS, 2011) to modulate protein interactions in in vitro reconstituted systems and in live cells.
Please click on the links below to read more about our explorations within the fields of GPCRs, kinases, and myosins.
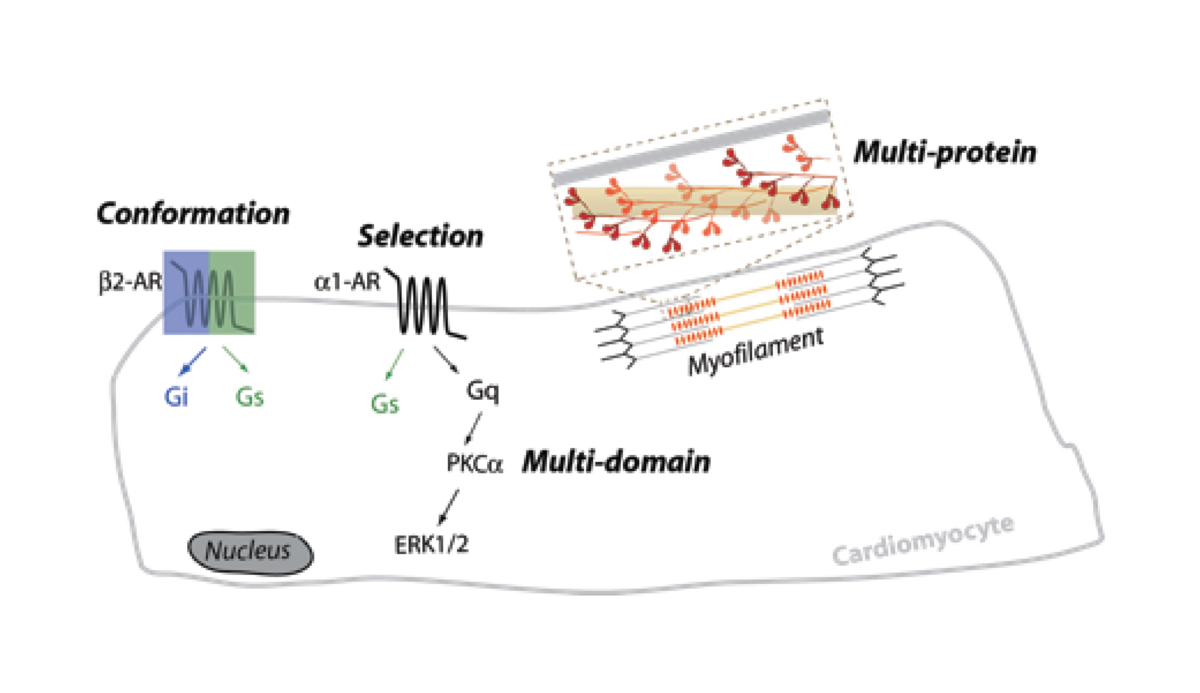
Figure 1: Integrated overview of lab research